I've carried out several projects analysing large and complex clinical datasets. Routinely those datasets are evaluated with in the first place, univariate and then multivariate logistic regression. However, I'm interested in applying the data-mining toolkit to understanding the datasets, including visualisation and applied machine learning models such as Support Vector Machines, Random Forests and Decision Trees.
Those methods often require more data, but the underlying algorithmics provides an alternative view on the dataset's inner structure, and thus alternative clinical algorithms. Such work always requires deep domain knowledge, or what follows, a tight-communication loop with the clinical collaborators.
I am currently leading efforts to standardize the existing electronic case report form (eCRF) for collecting data on patients with sexually transmitted infections (STIs), with a particular focus on Mpox. At present, there is no standardized eCRF specifically designed for this field.
Throughout these projects, I've developed a toolkit to perform the ML pipeline for any new dataset rapidly. I wanted to extend this to first compare the performance of the classifiers to the multivariate classification and logistic regression methods and then extend it to the alternative tree-based clinical algorithms.
All clinical studies rely on a data collection tool, which can range from a simple Excel database to more sophisticated platforms like REDCap. The content of the data capture tool—such as the variables and their definitions—establishes the framework for downstream data analysis. The encoding, including the ontology used, plays a crucial role in ensuring the FAIRness (Findability, Accessibility, Interoperability, and Reusability) of the resulting dataset. Across various projects, I have been actively engaged in structuring, harmonizing, and standardizing electronic case report forms (eCRFs) to promote consistency and data quality.
Research on post-COVID syndrome: current gaps and future perspectives
Learning from Post COVID-19 condition for epidemic preparedness: a variable catalogue for future Post-Acute Infection Syndromes
Orchestra is a large, international, interdisciplinary project funded by EU with almost 30 million euros - of which our group constitutes a coordination package. The call was opened in an emergency manner in March 2020 as a part of the EU response to the pandemic. I was engaged at the project-proposal writing stage, creating visualisations and reviewing sections related to data management.
Since the beginning of the project, October 2020, I've been devoting a considerable amount of my time to the Orchestra project. I am a representative of the WP1 to WP7 data-management package (Figure), helping with data flow and communication across the project.
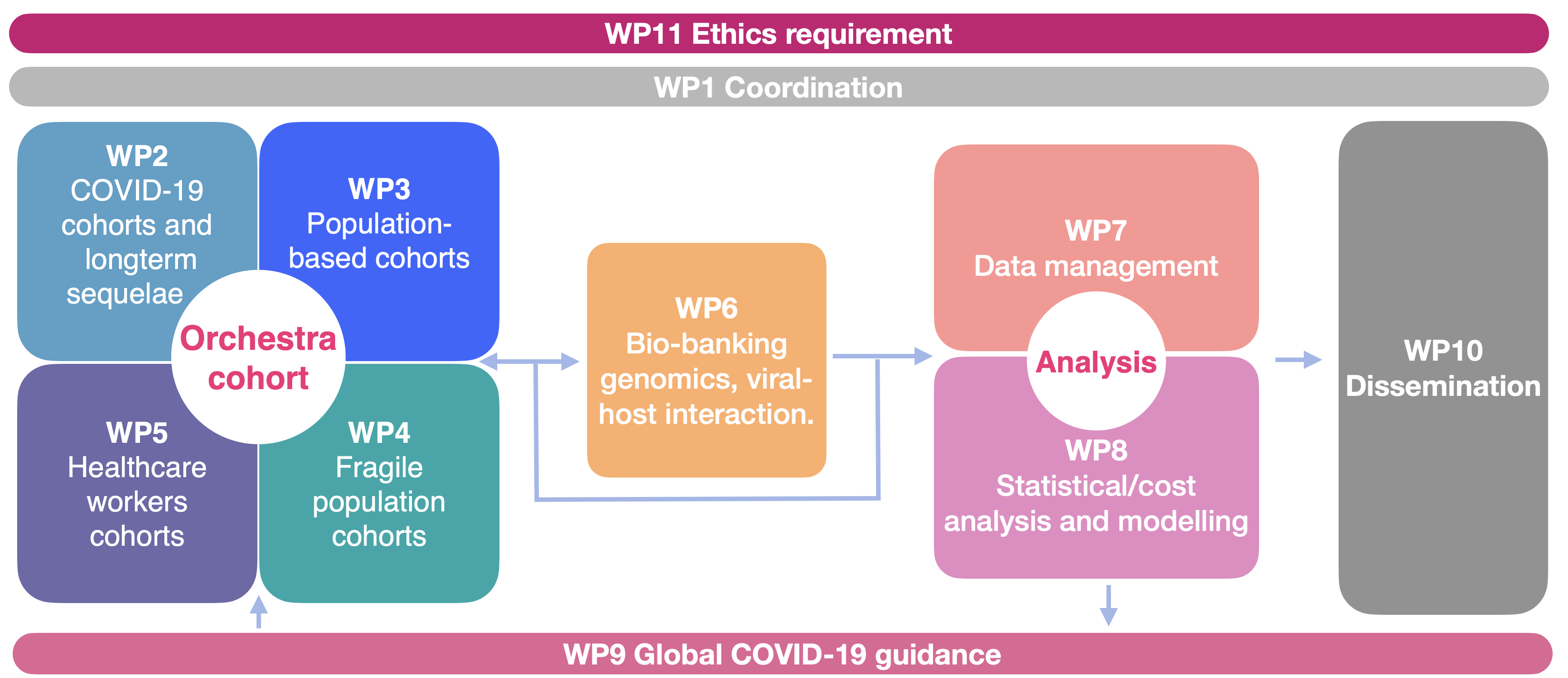
Within the ORCHESTRA project, my role involved managing the Post-COVID-19 working group, where I contributed to data management, quality control, and visualization. Additionally, I supported the work of WP6. Our publication in *Eurosurveillance* presents an in-depth bioinformatics analysis of SARS-CoV-2 sequences from infected patients, examining the in-patient evolution of viral strains. We demonstrated that viral infections are not limited to a single infecting strain but also give rise to subpopulations of viral sub-species.
Latent transition analysis for longitudinal studies of post-acute infection syndromes
Persistent Low-Grade Inflammation and Post-COVID Condition: Evidence from the ORCHESTRA Cohort
Quasi-species prevalence and clinical impact of evolving SARS-CoV-2 lineages in European COVID-19 cohorts, January 2020 to February 2022
Chemosensory assessment and impact on quality of life in neurosensorial cluster of the post COVID 19 syndrome
Clinical phenotypes and quality of life to define post-COVID-19 syndrome: a cluster analysis of the multinational, prospective ORCHESTRA cohort
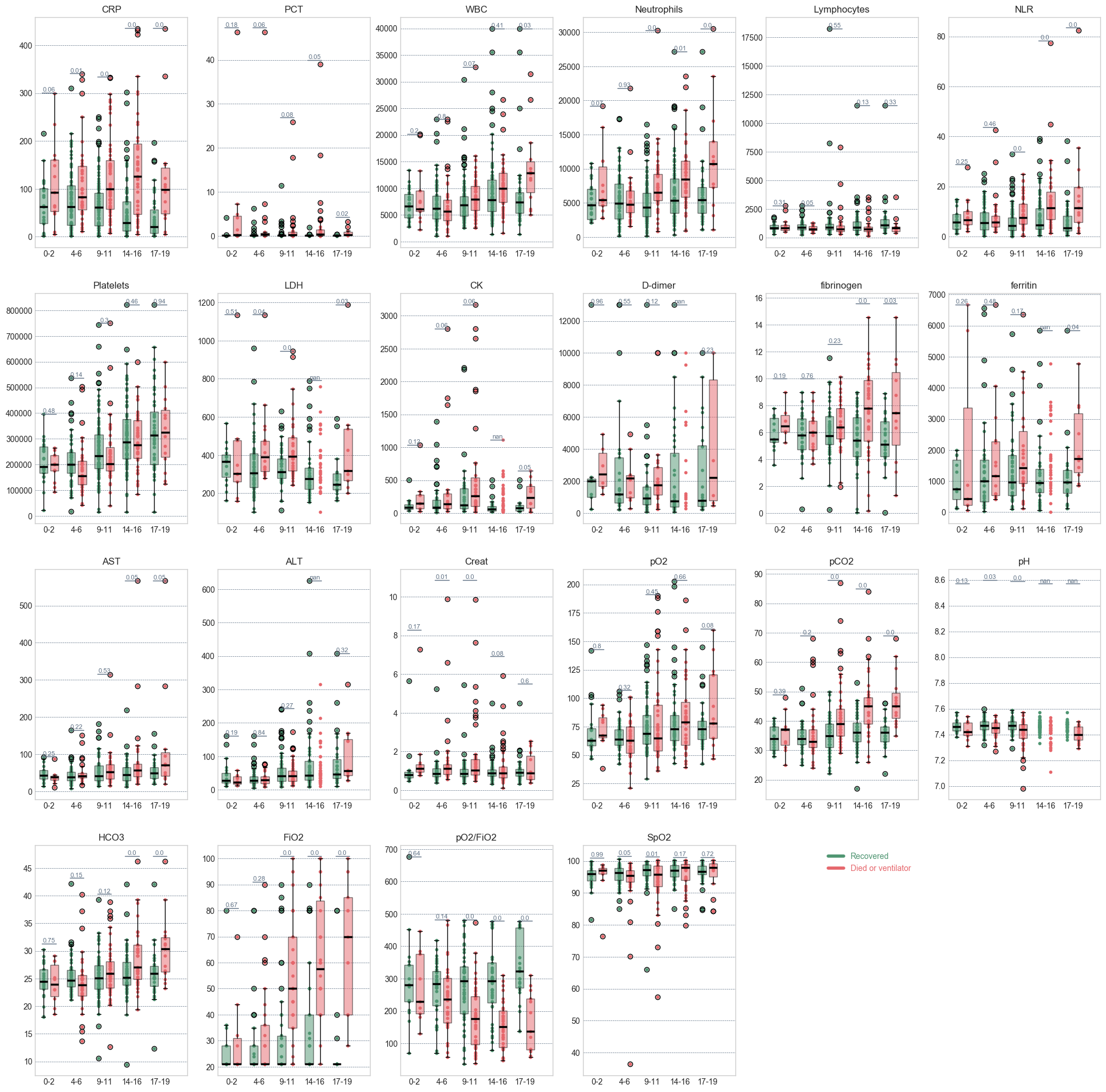
COVID19 pandemic added a lot of work to the collective plate of the Infectious Diseases department in Verona. North of Italy was one of the most affected regions in the first months of 2020. However, thanks to the experience, we quickly updated the data-collection tools to study the new virus. This dataset was too small to attempt a ML-based prediction, nevertheless we observed the development of the parameters for the positive/negative outcomes Figure.
Bloomy project was a multi-centre collaboration of the German hospital-clinics aiming to develop an as early as a possible scoring system for the progression of sepsis. The dataset included ~2,500 patients with sepsis. The study followed patients longitudinally, including a post-hospitalisation follow-up, with quality of life scores. The dataset had thousands of features describing patients' demographics and comorbidities, hospital stay, treatment, antibiotics and microbiological information such as infecting bacteria. The dataset was quite complex but riddled with missing values - that needed to be inputted.
We have approached it in two ways: analysing the regression coefficients and Machine Learning methods. I build sequential models at different time points in the treatment: 3rd, 7th, end of therapy timepoints, using mainly RF, SVMs and ADABoost models (Figure). Those were subsequenlty used to select variables for the Cox proportional hazards, logistic regression, and parametric survival regression.

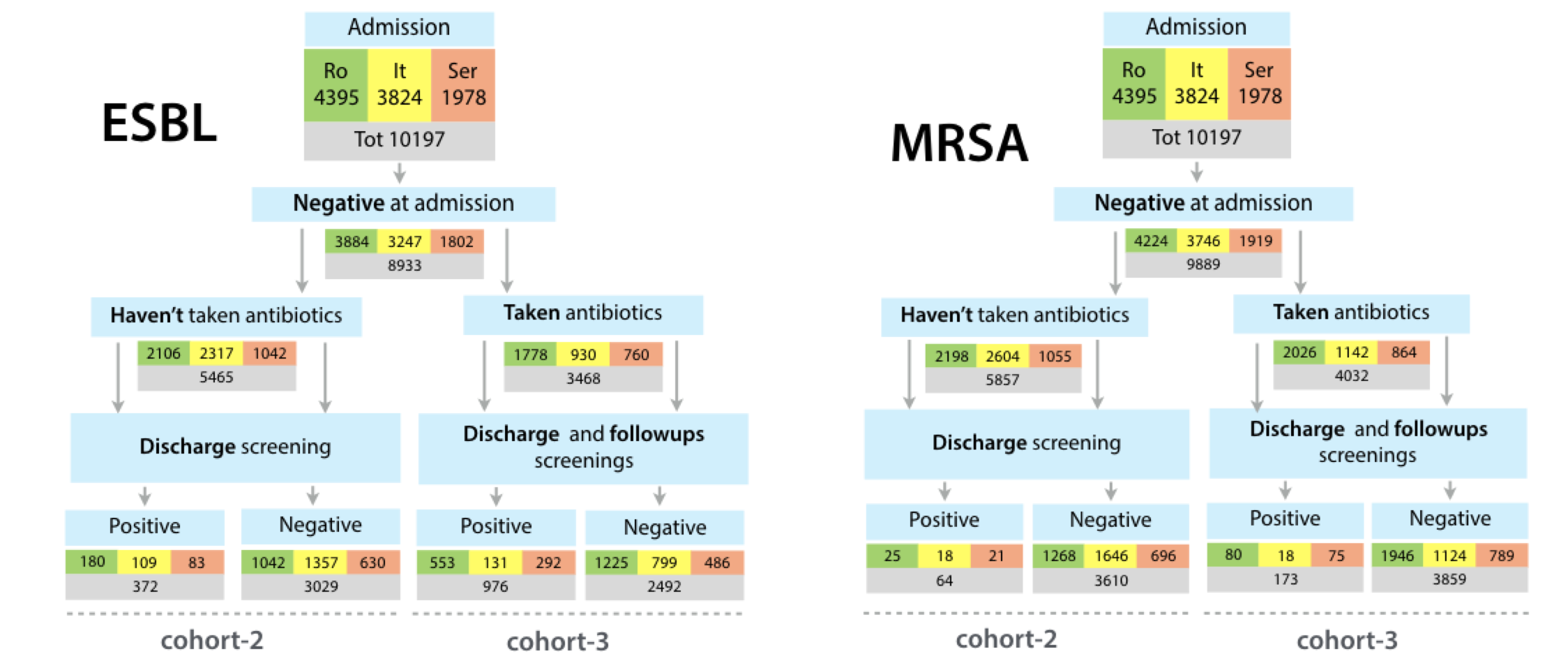
SATURN project investigated the relationship of antibiotic prescribing to the colonization with the ESBLs, and MRSA - two multi-drug resistant bacteria common in the nosocomial settings. The dataset had ~10,000 patients, whose data were collected in three countries. The participants were screened at admission to ensure their colonization happened during the study.
Figure shows basic results. As expected, many more patients got colonized with ESBLs than MRSAs. Among those patients who took antibiotics, the level of colonization with both ESBL and MRSA was ~3 times higher than in the respective group who weren't treated with antibiotics.
To analyse this dataset I've deployed a Mongodb instance to keep and quickly access the patients within the dataset. Next, I implemented a complex encoder to produce a numerical vector representing each patient, to be later fed to Random Forest, Ada Boosting, Gradient boosting and other machine learning methods. In order to encode often complex antibiotics therapy - I first included a simple length of treatment per antibiotic. For each antibiotic combination, several days, any two antibiotics were taken together. Finally, I included a binary vector that, again for each combination, encoded if one of the antibiotics was prescribed after the other. This way, a complete description of the antibiotic therapy is included in a patient vector. The 10,000 patients did not fully and equally capture the diversity of the antibiotics therapy. Nevertheless, machine learning methods were able to distinguish more and less selective antibiotic combinaitons.